Our Mission
Biotechnology is one of the most promising technologies when it comes to replacing energy-intensive or environmentally polluting processes with sustainable alternatives. But it is precisely the enormous complexity of biological systems that makes it so difficult to reliably apply this technology on an industrial scale. The production of raw materials or pharmaceuticals is typically biotechnologically optimized in high-throughput processes in which thousands of steps are carried out in parallel. The issue with these automated systems is that they are specialized in one biotechnological process only.
In this context, the goal of the KIWI-biolab is to develop self-learning robotic systems able to independently determine the optimal experimental conditions for certain biotechnological processes using large-scale database search. Assisted by machine learning methods, the system should also be able to improve these processes and develop new solutions. In addition, the system must be able to recognize image data, such as microscopic images or other optical signals, to autonomously monitor the production process and optimize it.
The development of such a system requires a close, interdisciplinary cooperation of diverse research fields, including computer science, engineering and biotechnology. The KIWI-biolab gathers together international experts, collaborators and partners from industry, who distribute their contributions along four task forces: Active Learning, Model Predictive Control, Signal Processing, and Automation.
TF1: Machine Learning for Bioprocess Forecasting and Control and Improved Enzyme Screening
This joint Task Force of TU Berlin and the University of Hildesheim is involved in several machine learning tasks within the KIWI-biolab, building on the experience of the ISMLL Hildesheim in time series forecasting, supervised learning for heterogeneous data and meta learning on the one hand, and the experience of the bioprocess group at TU Berlin with mechanistic modelling of bioprocesses on the other hand.
Task Force 1 is developing forecasting models for bioprocesses, given culture conditions, feeding profiles and metadata of processes and organisms. Traditionally, these processes have been modelled by estimating the parameters of biologically and biochemically motivated mechanistic models. We propose to use domain agnostic forecasting models alongside hybrid models that incorporate mechanistic components and mass balances.
An important goal of the Task Force is to model the forecasting problem across experiments and organisms, in order to transfer information from old experiments to new ones, thus decreasing the number of experiments required for the characterization of a new process. This can be seen as an instance of (Bayesian) meta learning.
Together with the bioprocess experts in the team, we will also investigate whether improved general forecasting models can be ‘distilled’ into improved mechanistic models.
One essential aspect of the modelling is active learning or sequential optimal experimental design. The purpose is to propose the most informative experiments to reduce the number and cost of experiments needed to achieve a certain forecasting quality. The active learning methods will have to compare to the established methods of (sequential) optimal experimental design for classical parametrized models.
The ultimate goal of the process forecasting is to enable the optimization of culture conditions and feeding profiles with respect to maximal yield of some product. In collaboration with Task Force 2, we will use general and hybrid forecasting models for model predictive control, but also work on a reinforcement-learning based control mechanism in end-to-end learning.
Together with the University of Greifswald, Task Force 1 is working on methods for an improved enzyme screening. In a ‘directed evolution’ scenario, we want to go beyond the selection of best variants towards the recommendation of new variants by means of machine learning models, using previous screening, sequence information and possibly structural information. The open-source and free software LARA suite, developed at the University of Greifswald for automated high-throughput screening, is to be extended in order to facilitate the integration of machine learning models (as developed in the KIWI-biolab) into the screening workflow.
TF2: Model Predictive Control for High Throughput Bioprocess Development
Bioprocess development is rapidly accelerating pushed by the integration of advanced liquid handling stations with embedded parallel minibioreactors into modern laboratories. Nevertheless, the complexity of these parallel processes demands model-based methods for process design, optimization, scale-up, and scale-down to exploit its full capacity.
Even though some applications have been reported in parallel mini-bioreactors (Abt 2008), further development is needed to achieve a fully autonomous operation for up to 48 parallel dynamical experiments in a process-wide design and optimization scheme. This Task Force will deliver the backbone for online State and Parameter Estimation (SPE), nonlinear Model Predictive Control (nMPC), and online sustainable process optimization.
The State and PE challenges will be tackled using a robust MHE for complex nonlinear systems with event based simulations, appropriate estimation methods (e.g. Maximum Likelihood) and statistical error models, and advanced regularization techniques for handling large parameter spaces.
The nMPC framework, including model free methods (e.g. reinforcement learning (Kim 2020)), hybrid models (e.g. Step-Wise Gaussian Process models (Bradford 2018), symbolic regression models), and first principle dynamical models (e.g. macrokinetic growth models (Anane 2017)), will be directly coupled with the DataBase and low level automation in TF4.
Optimization techniques that shall enforce autonomous and parallel bioprocess development to search for the most sustainable production strategy will be screened, further developed, and integrated. To this aim proper relaxations of large optimization problems with sustainable cost functions (e.g. planetary boundaries (Algunaibet 2019)) in parallel structures need to be developed to cope with the computational efficiency required in the nMPC framework of the 48 minibioreactor facility (Krausch 2019).
Finally, this implementation will be highly modular (running in micro systems) and fully compatible with the tools and programs developed in Task Force 1 (ML models for forecast, active learning, and optimization) and Task Force 3 (automated signal processing tools) so as to enable a systematic computation, analysis, and evaluation of corresponding large and complex optimization problems (Martinez 2013), and seamless integration of all elements developed by the KIWI-biolab and partners in one workflow.
References:
Abt 2008 – Abt, Vinzenz, et al. “Model-based tools for optimal experiments in bioprocess engineering.” Current opinion in chemical engineering 22 (2018): 244-252.
Algunaibet 2019 – Algunaibet, Ibrahim M., et al. “Powering sustainable development within planetary boundaries.” Energy & environmental science 12.6 (2019): 1890-1900.
Anane 2017 – Anane, Emmanuel; Neubauer, Peter; and Cruz Bournazou, M. Nicolas. “Modelling overflow metabolism in Escherichia coli by acetate cycling.” Biochemical Engineering Journal 125 (2017): 23-30.
Bradford 2018 – Bradford, Eric, et al. “Dynamic modeling and optimization of sustainable algal production with uncertainty using multivariate Gaussian processes.” Computers & Chemical Engineering 118 (2018): 143-158.
Kim 2020 – Kim, Jong Woo, et al. “A model-based deep reinforcement learning method applied to finite-horizon optimal control of nonlinear control-affine system.” Journal of Process Control 87 (2020): 166-178.
Krausch 2019 – Krausch, Niels, et al. “From Screening to Production: a Holistic Approach of High-throughput Model-based Screening for Recombinant Protein Production.” Computer Aided Chemical Engineering. Vol. 48. Elsevier, 2020. 1723-1728.
Martinez 2013 – Martínez, Ernesto C.; Cristaldi, Mariano D.; and Grau, Ricardo J. “Dynamic optimization of bioreactors using probabilistic tendency models and Bayesian active learning.” Computers & Chemical Engineering 49 (2013): 37-49.
TF3: Advanced Signal Processing
The goal of Task Force 3 is to investigate how signal processing and visual computing methods can aid biomolecular processes through a fast online analysis of large amounts of data being generated in parallel platforms. A machine learning layer for data analysis and smart sensor development will select the analysis methods to be used, calibrate them, and assess its implementation for online operation. The different sensors, probes, enzymatic analyzes and microscopic images deliver noisy data. Tools processing the data and extracting valuable information are essential to maximize the knowledge gained from each experiment.
To this aim, the following methods will be implemented:
- Blind Signal Separation (BSS) for complex spectral data with high background noise and low concentration metabolites (mL-μL) in the cultivations.
- Spectral unmixing by (Bayesian) non-negative matrix factorization and matrix completion, where spectra of components may be partially known.
- Supervised learning of relevant properties of spectra/images from annotated data using deep learning.
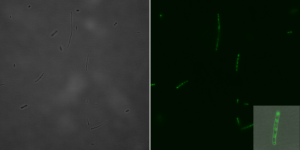
Bright field (left) and fluorescence microscopic (right) image with a 1:5 magnification of a single E.coli cell 135 min after induction with IPTG. Image: Irmgard Schäffl.
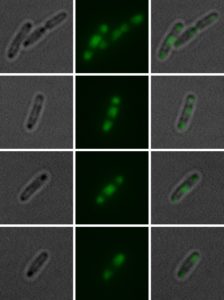
Bright field (left) and fluorescence microscopic (middle) image with a 1:5 magnification of a single cell 45 min after cultivation start. The panels on the right show a layered bright field and fluorescence image. Cells exhibit a “triple” structure of inclusion bodies (two at polar region, one in the middle). Image: Irmgard Schäffl.
TF4: Robot Operation and Control
Our team is responsible for operation and control of the High Throughput laboratories. That implies the data generation for feeding the models to be developed in Task Forces 1 and 2, and the experimental implementation and application of their artificial intelligence-based approaches, as well as the analytical strategies developed in Task Force 3.
The robotic facilities of the KIWI-biolab must be able to autonomously perform the experiments as designed by higher level applications, programs and, therefore, develop a robust and efficient process. The optimization strategies proposed by the machine learning artificial intelligence algorithms represent a major challenge for the control of the robotic facility. Effective and fast re-scheduling planning of resources and analysis methods is required to ensure robust operation under changing strategies.
The Model Predictive Control (MPC) strategy will be directly embedded to the dynamical machine learning algorithms developed in Task Forces 1 and 2, building an advanced active learning framework. Convolution Neural Networks algorithms on strictly continuous differentiable functions will be developed to describe the complex dynamics of the biological processes.
The structure, connection and data architecture developed in here creates the basis for the KIWI-biolab project and its success. In order to give the algorithms maximum freedom for optimization, a constant rescheduling of the experiment is necessary, including the automated process and the inputs. At the same time, the efficiency, robustness and traceability of the experiments must be ensured. For this challenge, we will use AI algorithms to address the scheduling problem. The automated data processing, which serves as the basis for the tools in the KIWI-biolab, will be expanded. The recorded data will undergo an automated data pre-treatment step (outliers, correlated parameters, etc.). In order to be able to master the high-dimensional process with up to 500 variables to be monitored, the process data processing, visualization and storage will be developed in close coordination with the subcontractors.
The experimental work takes place in automated high-throughput laboratories at the TU Berlin and at the University of Greifswald. The test planning, test execution and test evaluation will meet the F.A.I.R data principles. The aim is to create a F.A.I.R data pipeline by generating and using open (source) data and communication standards.
To enable a standardized experimental framework SiLA standards will be implemented for controlling and coupling the laboratory instruments. Uniform data storage and processing will be reached via the AnIML standard. The standardized and automated biotechnology laboratory enables interoperative work. This ensures that the achievements of this project can be transferred directly to external laboratories and bioprocess development projects.
